Full company details
Mad City Labs Inc.
 2524 Todd Dr.
2524 Todd Dr.
Madison, WI 53713
United States
Phone: +1 608-298-0855
Fax: +1 608-298-9525
Precision Positioning Aids Microscopes with Tracking Viral Progression
BioPhotonics
Jul/Aug 2023Imaging fluorescence at the single-particle level provides nanopositioning systems with the spatial resolution required to track viruses as they enter the cell and replicate.JENICE CON FOO, MAD CITY LABS, INC
The rapid spread of COVID-19 and other diseases has shown that understanding viral infection is critical to the health of people around the world. In particular, understanding the dynamics of the earliest stages of infection is important for developing prevention strategies, while understanding replication and late-stage processes are important for developing medical, pharmaceutical, and public health interventions. Human virus particles are typically between 20 and 200 nm in diameter, and so their visibility is just beyond the diffraction limit of optical microscopy. Not only do they come in varying sizes, but they also appear in a range of shapes and form factors. For many reasons, it is preferable to study single particles rather than conducting ensemble measurements, and high-precision positioning can provide the high spatial resolution that is required for this type of single-particle experiment.
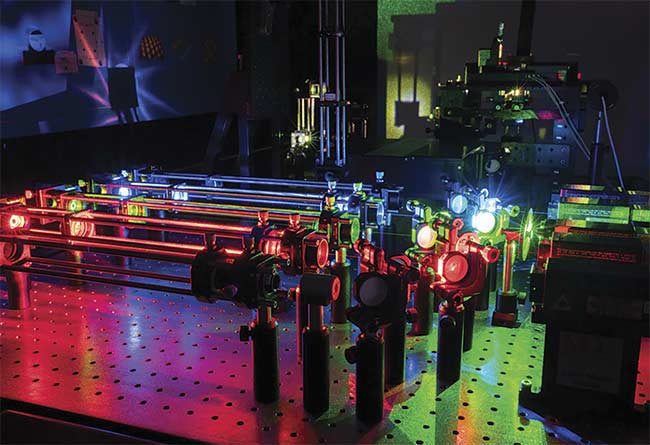
A multicolor single-molecule microscope with the RM21 Micromirror TIRF microscope in the top right of the photo. Courtesy of Mad City Labs, Inc.
This article discusses two different techniques for studying virus particles, which rely on this high-precision positioning in microscopic research.
Viral infection process
Viruses are nucleic acid (DNA or RNA) surrounded by a protein shell, or capsid. Some viruses have a lipid membrane surrounding the capsid, called enveloped virus particles. A virus cannot replicate alone — it needs to “hijack” mechanisms of a host cell to replicate. Given the minute size of the typical virus particle that is just beyond the diffraction limit, imaging systems enabled by nanopositioners are needed to follow its progression.
All of us have been subjected to a viral infection and are broadly familiar with how infections occur. However, to understand how you can study single-virus particles, it is useful to think about viral infection as a four-step process: entry, intracellular transport, replication, and exit — which results in the release of new viruses. Each of these steps is of interest to understand how to mitigate the effects of viral infections (Figure 1).
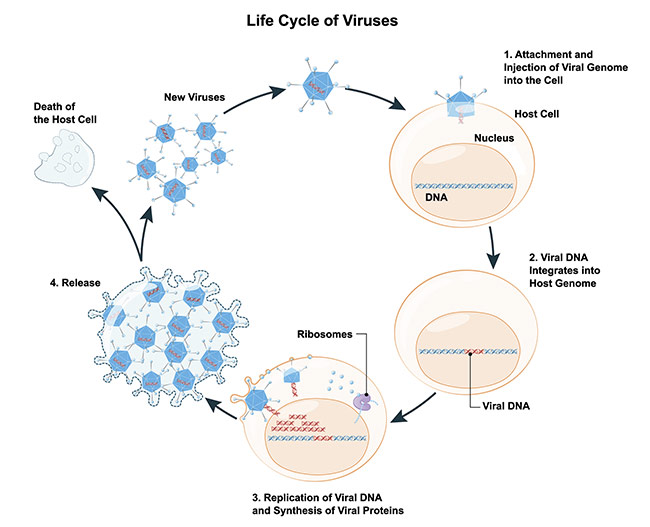
Figure 1. Schematic of the life cycle of a virus. Courtesy of iStock.com/ttsz.
The first step in viral infection, cellular entry, is a confluence of multiple related steps. Since viruses cannot replicate alone and need a host, this step is the subject of intensive study by researchers and clinicians. This step comprises proximity and attachment mechanisms — how a virus particle finds and lands on a host cell — as well as the actual process of entry into the host cell. One of two mechanisms is used for entry: membrane fusion (enveloped viruses) or endocytosis (enveloped or nonenveloped viruses). The knowledge of how the process allows the virus to enter a cell can yield insights into clinical and pharmaceutical interventions that are required to mitigate viral infections.
Once the infection enters the host, the next steps of viral infection are intracellular transport and replication. In other words, the virus particle needs to arrive at a suitable site to exploit the host cell resources for replication. There are several ways for viruses to be transported within the cell, but ultimately the goal for a virus is to reach the replication site, such as the cell nucleus. Replication strategies vary among virus families and therefore the exploitation of the host cell resources also varies.
The final step is shedding or exiting the host cell; this essentially releases the replicated virus particles to propagate the viral infection. Broadly speaking, virus particles are released either via cell lysis of the infected cell (generally nonenveloped viruses) or exocytosis (enveloped viruses).
Challenges in imaging
While the four-step breakdown of the infection process appears simple in theory, each step is divided into many intermediate processes that vary in complexity when researchers attempt to distinguish them. The challenge lies in finding a relatively simple part of the process on which to focus the imaging technology. Cell entry is an ideal step to approach with microscopy: This step starts with a known virus particle and a known host cell, so the priority at this stage is understanding the underlying mechanisms of entry.
Vital questions about viral infection, with implications for public health and medical interventions, occur during the entry step: Does the shape of the virus particle matter? Why do some virus particles enter the host and not others? What are the factors, structural or chemical, that determine landing areas or affinities for viruses on host cells?
As noted, the size of virus particles is beyond the diffraction limit of light. So how can they be seen? Single-molecule localization microscopy (SMLM) is well suited to address this question.
For SMLM, components of the virus particle can be labeled with fluorophores. Fluorophores may be organic dyes, fluorescent proteins, or quantum dots and can be photoactivated (via a laser) or chemically activated. The fluorophore is imaged through the microscope and appears on the camera as a spot with a defined width known as the point spread function. SMLM benefits from the fact that the location of a single fluorophore can be determined with high precision if their point spread functions do not overlap. It is important to note that localization precision is primarily limited by signal-to-noise ratio (SNR) rather than light wavelength or imaging pixel size. The SNR can be materially affected by selectively illuminating the sample, fluorophore activation, and low-noise nanometer precision positioning to ensure the best localization statistics.
In the Ivanovic Lab at the National Institute of Allergy and Infectious Diseases, researchers are using single-molecule micromirror total internal reflection fluorescence (TIRF) microscopy to study cell entry mechanisms and describe the relationship between virus particle structure and the early steps of infection.
TIRF microscopy is based on the principle that when light encounters the interface between media with different refractive indices, the light can be refracted or reflected depending on the angle of incidence. At a specific critical angle, the light is entirely reflected from the interface and an evanescent wave is generated in the lower refractive index media. The evanescent wave exponentially decays with distance from the interface; therefore TIRF penetration depth is between 30 and 300 nm, which leads to a very selective illumination of the sample.
In the case of micromirror TIRF, the excitation light enters and exits through the microscope objective via micromirrors. Employing multiple excitation wavelengths enables the use of micromirrors instead of a dichroic mirror and leads to improved SNR simply because the light reflected off the mirrors is wavelength independent and the excitation and emission beam pathways are spatially separated.
Importance of positioning
The ability to perform multicolor imaging is an essential tool in answering these questions about cell entry, such as the impact of particle size and shape, membrane fusion mechanics, and virus assembly and structure on the entry process. The Ivanovic Lab TIRF microscopy approach includes high-resolution nanopositioners, which allow tight positional control. This is an important component in a microscopy system when imaging fluorophores that may be separated by only a few nanometers.
Researchers in the lab have been able to dissect and see intermediate components of the cell entry process, while simultaneously quantifying structural features of virus particles. For example, they have been able to identify a population of virus particles as pleomorphic — particles varying in shape and size. If a fluorophore is attached to a virus particle such that it scales with particle size, information can be garnered regarding particle size at a per-particle level, and in turn discover how particle size affects these cell entry processes.
It is also possible to assess virus assembly and disassembly using simultaneous multicolor imaging. The Ivanovic Lab has successfully labeled a virus core particle with one fluorophore and labeled an outer capsid molecule with a different fluorophore. A secondary fluorescence is detected when these two components come together, allowing the virus assembly to be visible in real time.
In Figure 2, the moment of cell entry is studied using SMLM. But what would be the mechanism to glean information regarding how viruses navigate the extracellular space? To understand how viruses reach their hosts, single-virus tracking is required.
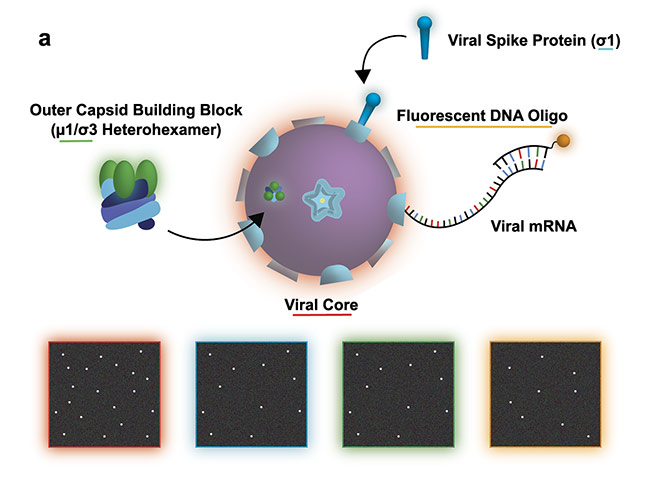
Figure 2a. Study of a reovirus assembly, disassembly, and transcription. The illustration shows the experiment design with the bottom four panels illustrating a possible view of the transcription/assembly for the labeled components: viral core (red), viral spike protein (blue), outer capsid (green), viral messenger RNA (mRNA) (orange). Courtesy of Ivanovic Lab/National Institutes of Health.

Figure 2b. Experimental data set demonstrating efficient two-color assembly on the microscope slide. Courtesy of Ivanovic Lab/National Institutes of Health.
High-speed tracking
The key challenge of particle tracking concerns observing a small object that is moving very fast. Recently the Welsher group at Duke University developed a novel 4D microscopy method based on the root technology of active feedback tracking microscopy.
As is the case with SMLM, the virus particle is labeled with a fluorophore to enable imaging and diffusing in 3D. The virus particle has a finite number of photons and can only be imaged in the focal plane.
The resulting instrument comprises two key elements. The first is a microscope called 3D-SMART
1, which performs active feedback tracking. A laser spot is rapidly scanned in 3D to “dance” along a specific path around the virus particle, causing it to emit photons. The photons are collected at a high speed, and their arrival times determine the location of the diffusing virus particle. When it is used alone, the virus particle can be tracked at a high speed but does not provide context related to the bodies and particles around it.
To facilitate the high-speed tracking, a high-speed three-axis nanopositioning system is employed. The photons emitted by the virus particle allow observers to estimate the position of the particle within a small region. The nanopositioner allows the user to move the entire sample to fix the particle in place for observation. Therefore, as the virus particle moves, the nanopositioner uses active feedback from the tracking to reposition the sample in real time for imaging purposes, keeping the virus particle centered in the field of view.
The instrument’s second element is a volumetric imaging microscope, enabling a technique called 3D-FASTR
2 built around the 3D-SMART microscope. 3D-FASTR is similar to 3D-SMART; a laser is scanned over the volume but over a much larger range. The purpose here is not to determine the position of the particle, but rather the “global” image for contextualizing the virus trajectory (Figure 3).
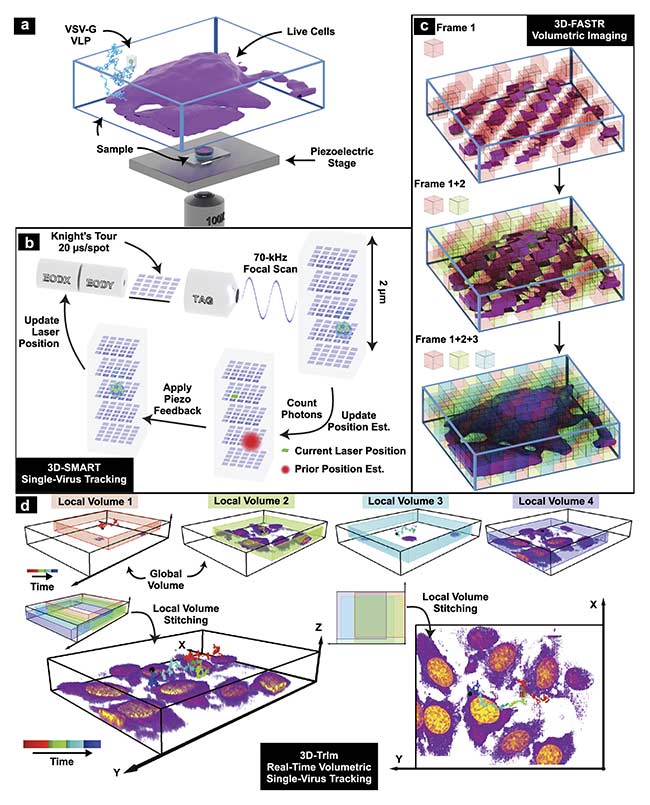
Figure 3. 3D tracking and imaging (3D-TrIm) in an experimental setup. Fluorescently labeled virus-like particles (VLPs) are added to live cells plated on a
coverslip. The sample is placed on a heated sample holder mounted on a piezoelectric stage (a). Overview of 3D-SMART tracking of single viruses. The electro-optic deflector (EOD) and tunable acoustic gradient (TAG) lens rapidly scan the local particle area. Photon arrival times and the current laser position are used to calculate the position of the virus within the scan area. Using the measured position, the piezoelectric stage moves to recenter the virus within the scan area (b). Concept of 3D-FASTR volumetric imaging. By outfitting a traditional two-photon laser scanning microscopy with an electrically tunable lens, a repeatable,
tessellated 3D sampling pattern can be generated during each frame time. Over a set number of frame times, the entire volume is sampled (c). Construction of global volumes in 3D-TrIm. As the virus diffuses, 3D-SMART moves the sample and the 3D-FASTR imaging system collects sequential volumes from different areas around the particle. These time-resolved local volumes can be used to generate an integrated global volume (d). Courtesy of Welsher Lab/Duke University.
Combining 3D-FASTR and 3D-SMART into a single instrument enables researchers to study virus particles in proximity to the host cell as well as the cell entry processes. And since virus particles tend to be heterogeneous, this combination of instruments can help investigate whether there are specific factors that impact the virus particles’ attachment to host cells.
The mechanics of infection
High-precision positioning has a role to play in a variety of microscopy applications as the world tries to grapple with the prospect of another pandemic. As single-molecule microscopy methods arise, the characteristics of high-resolution positioners become more important as researchers race to learn how the viruses work and thrive. Low noise and high resolution are fundamental characteristics of nanopositioners and are instrumental in surmounting some of the challenges regarding single-molecule microscopy and the tracking that has impeded earlier iterations of optical technology.
Methods such as colocalization single-molecule microscopy and single-virus particle tracking provide fundamental insights into the mechanics of infection. The importance of these insights became glaringly obvious as COVID-19 spread around the world and messenger RNA (mRNA)-based therapeutics advanced rapidly. Without the insights that enable these developments, it is difficult if not impossible to initiate pharmaceutical, clinical, and public health measures to mitigate the effects of viral outbreaks.
Meet the author
Jenice Con Foo is a member of the technical sales and marketing team at Mad City Labs, Inc. She obtained a Ph.D. in physics from La Trobe University Australia, followed by positions at the Synchrotron Radiation Center and the University of Wisconsin-Madison. Her research background is in instrumentation development; email:
[email protected].
References
1. S. Hou et al. (2020). Real-time 3D single molecule tracking.
Nat Commun, Vol. 11, No. 1, p. 3607.
2. C. Johnson et al. (2019). Continuous focal translation enhances rate of point-scan volumetric microscopy.
Opt Express,
Vol. 27, No. 25, pp. 36241-36258.