Mark Roskey, Caliper Life Sciences Inc.
Microfluidics and optical imaging are bridging the productivity gap in drug development.
The pharmaceutical industry is under intense
pressure to improve the efficiency of drug discovery and development. Despite continued
increases in research and development spending, the number of new molecular entities
receiving FDA approval as marketable drugs has decreased.
There are many reasons why pharmaceutical companies
are experiencing this “productivity gap,” but at least part of the problem
is that spending on discovery tools and methods has focused most heavily on high-throughput,
low-cost in vitro experimentation for early-stage “hit” identification.
The failure to link early in vitro experimentation with accurate, effective in vivo
testing causes drug attrition and inefficiency.
Experimental approaches that can link
high-quality results obtained in vitro with accurate and highly relevant in vivo
testing are being implemented in a number of drug discovery programs. This linkage
is starting to directly close the productivity gap. In March, the FDA issued a report
on issues facing the pharmaceutical industry and specifically identified areas of
opportunity that require better evaluation tools. These areas include qualifying
new biomarkers, developing new imaging technologies and improving the predictability
of human response from disease models.
Better in vitro testing
The traditional approach to in vitro testing —
combining advances in laboratory automation, combinatorial chemistry and genomics
— has enabled researchers to screen a very large number of candidate compounds
(well over 100,000 per day) in a relatively simple activity assay. This generates
many low-quality screening hits that typically require significant resources to
follow up.
The hits that come from this high-throughput
approach have been plagued by significant numbers of false-positives and false-negatives
and have, on the whole, been found to exhibit low pharmaceutical relevance —
generally having a very high attrition rate as the molecules are taken from early-stage
hits through development of lead candidates. The problems of low data quality and
lack of relevance can extend into the lead optimization stage of development. Researchers
need better ways to test new lead compounds in high-quality cell-based assays to
obtain as much information on side effects and relevancy as early in the process
as possible.
Microfluidic technologies can have
a significant impact on the generation of early-stage high quality data by providing
a high-throughput platform technology that can more accurately measure enzyme activity.
Microfluidics provides a closed controlled experimental environment that enables
precise measurement of the specific activity of an enzyme.
Typical experiments in this format
generate assay data with very high precision (Z’ <0.9) and very low coefficients
of variation (<5 percent). These parameters enable identification of compounds
that are inhibiting an enzyme at the 10 percent level, which provides critical information
for compound development and for understanding potential side effects. Furthermore,
because the standard microfluidic assay separates substrates from products, it is
not subject to fluorescent artifacts that typically compromise homogeneous assays.
Microfluidics-based assays can be applied
to a wide variety of drug discovery target classes, including kinases, phosphatases,
proteases, lipid-modifying enzymes, histone deacylases and other specific enzymatic
targets, allowing very accurate measurement of the conversion of a substrate to
a product. The technology also enables real-time, high-throughput determination
of enzymatic activity parameters and the accurate determination of the inhibition
coefficient, leading to direct determination of the compound’s inhibitory
mechanism of action. Additionally, these assays can profile a hit compound against
a variety of similar targets to rapidly assess cross-reactivity and to identify
potential downstream side-effect issues.
In vivo imaging
As compounds move from the hit stage into lead
optimization, testing hits for their functional efficacy in vivo becomes critical.
Optical imaging techniques are increasingly helping to improve pharmaceutical relevance
by enabling researchers to “see” whether candidate compounds are appropriately
affecting the functional targeted biology in an experimental animal model.
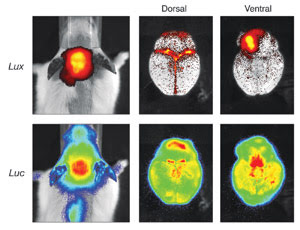
In vivo imaging of an infectious disease and neuronal damage mouse model is useful
for development of antibacterial drugs. The top panel shows the luminescent signal
generated from bacterial luciferase (Lux) and indicates the presence of infectious
organisms. The bottom panel is the signal from firefly luciferase (Luc) in this
mouse and indicates neuronal damage as a result of the bacterial infection.
Using in vivo optical imaging, a single
animal can be imaged over multiple days throughout a drug evaluation study rather
than being sacrificed each time a measurement is made — as is required using
traditional physical measurement or histological-based methods. This improves data
quality by reducing the variability of measurements from the same animal over time,
and it substantially reduces the number of animals needed for a particular study.
The technology also simplifies preclinical compound dosage studies and is an ideal
way to obtain more relevant pharmacokinetic and pharmacodynamic data.
Optical imaging is made possible by
labeling genes, cells, bacteria and laboratory animals (predominantly mice and rats)
with a luciferase reporter molecule or a biologically specific fluorescent probe.
The light from these reporter molecules or probes can be accurately quantified using
sensitive imaging instruments that provide three-dimensional information and enable
the researcher to observe the functional biology.
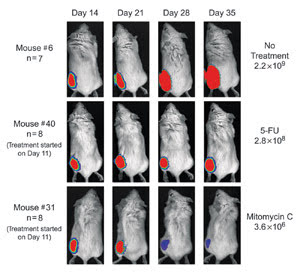
Luciferase labeled PC-3M tumor cells were injected into mice. Mouse #6 received no treatment,
and the tumor grew from day 14 through day 35. Mouse #40 was treated with the chemotherapeutic
agent 5-Fluorouracil (5-FU). Mouse #31 was treated with the agent mitomycin C and
showed significant reduction in tumor growth.
Light output can be directly correlated
with the number of labeled cells or the level of expression of a particular gene.
A typical in vivo drug analysis experiment may involve injecting tumorogenic cells
labeled with luciferase directly into a small cohort of mice and then following
the progress and spread of those cells both with and without drug treatment. Tracking
tumor formation by monitoring the level of light production from the cells gives
a more accurate picture of tumor growth and metastasis because the light signal
correlates more specifically with the number of active cells than physical measurement
of tumor size. The technique is applicable to a wide variety of cancer cell types
and oncogenic models.
Optical imaging is also ideal for drug
discovery in infectious disease applications because bacteria can be easily labeled
with luciferase and tracked quantitatively in mice to look at the specific effects
of antibacterial compounds.
Besides labeled tumor cell lines, light-producing
transgenic animals have been developed for specific areas such as central nervous
system and stem cell research, metabolic disease and cardiovascular applications.
For example, an important model for
the central nervous system is the glial fibrillary acidic protein light-producing
transgenic animal, which generates a specific signal in response to neuronal damage.
Drug discovery researchers working on a variety of disorders related to the central
nervous system, including Alzheimer’s disease, multiple sclerosis and Parkinson’s
disease, can use this model to directly visualize neuronal damage over time.
A similar example in metabolic disease
research is the availability of specific light-producing transgenic animals that
report on the induction of insulin. This animal model can be used to improve the
relevance of diabetes drug discovery programs by generating a quantitative signal
upon induction of insulin.
Many new drug development programs
are targeting inflammation, which plays an important role in many forms of cancers
and can be a significant component of cardiovascular disease. Optical imaging can
improve the relevance of lead candidates in these therapeutic areas by being used
to evaluate compounds in vivo. Several light-producing transgenic animal models
are available, such as those that report on the regulation of critical genes in
well-known inflammation pathways. Early-stage compound evaluation in these models
produces a rapid readout of how a compound specifically interacts with the actual
pathway in vivo and provides an early indication of compound efficacy while providing
some indication of potential safety issues.
Bridging the gap
In many drug discovery programs, the tools for
in vitro compound identification have been developed in centralized high-throughput
screening labs. Technologies used for downstream in vivo evaluation experiments
are decentralized and typically reside in the therapeutic groups. Currently, there
is no direct linkage between in vitro tools and animal studies. Significant improvement
in the overall process and the speed at which relevant compounds can be developed
for clinical trial can be enabled by linkage of researchers, processes, tools and
technologies from in vitro assays through in vivo profiling.
A key example of how both microfluidics
and optical imaging can accelerate the drug development process can be found in
kinase research programs focused on the identification of new oncology drugs. By
using microfluidics in a primary and secondary screening program, researchers can
generate the highest-quality data on the specific characteristics of an inhibitory
molecule. For example, several groups have identified specific inhibitors for IkappaB
kinase, an oncology and inflammation-related kinase target, using this approach.
Microfludic-based systems can then profile the lead inhibitor against many related
kinases for an indication of potential side effects.
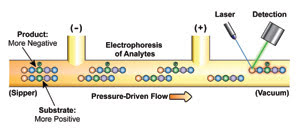
Microfludic separation of the product and substrate from
a kinase inhibition assay is shown. High-throughput quantitation of the substrate
and product of the reaction contributes to the high quality of the kinase assay.
Once a solid candidate has been identified
in this in vitro enzymatic assay, cell-based reporter gene assays can be developed
to gain quality data on the compound’s ability to enter the cell, on its
cytotoxicity and on its ability to affect the kinase in a cellular environment.
Next, cell lines that report on the activation of that specific kinase in a pathway
can be generated and profiled rapidly, again using microfluidics, to gain insights
into the effect that the kinase inhibitor is having on the specific pathway.
These pathway-reporting cell lines
can then be used as key reagents for the in vivo testing of an inhibitor. The cell
lines can be injected into mice, and the cells’ ability to form tumors and
to metastasize can be followed and quantified using optical imaging. Representative
sets of these mice can then be treated with the lead inhibitor compounds, and the
in vivo pharmacology of the compound rapidly assessed. This type of experimental
approach was used in the development of the multikinase kinase inhibitor Sutent,
which was fast-tracked and received FDA approval early this year. This case demonstrates
the power of bridging in vitro and in vivo experimentation to significantly influence
development time lines.
Data quality and pharmacological relevance
can be directly affected by better tools and by more relevant methods. The linkage
of early-stage high-quality hit identification tools with best-in-class technologies
for optical imaging is clearly having a significant impact on improving pharmaceutical
development productivity.
Meet the author
Mark Roskey is vice president of reagents and
applied biology at Caliper Life Sciences Inc. in Hopkinton, Mass.; e-mail [email protected].