A new multispectral device is said to have produced the largest microscope image ever, combining 13 color channels gathered by thousands of microlenses into a nearly 17-gigapixel picture.
Able to rapidly process very large amounts of biomedical imaging data, the system addresses what has been a major bottleneck in pharmaceutical development, according to a team of researchers from the U.S. and Australia.
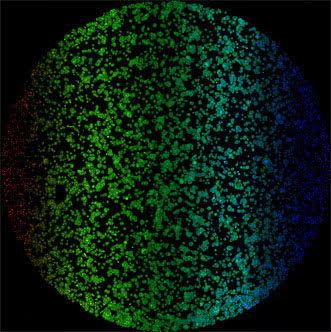
Parallelized multispectral imaging. Each rainbow-colored bar is the fluorescent spectrum from a discrete point in a cell culture. The gigapixel multispectral microscope records nearly a million such spectra every second. Images courtesy of Optica.
"Pharmaceutical research is awash with cutting-edge equipment that tries to image what is happening at the cellular level and smaller," said Antony Orth, a former Harvard University researcher now at RMIT University in Melbourne, Australia. "We recognized that the microscopy part of the drug development pipeline was much slower than it could be and designed a system specifically for this task."
Multispectral imaging is used for many types of medical research and not only produces an image but also provides data about the specific colors within that image. Researchers are able to study the frequencies to learn about the composition of biological samples and chemical processes taking place within them. Observing how cells and tissues respond to specific chemicals and experimental drugs is essential for pharmaceutical research, particularly for cancer treatment.
To demonstrate their design, the researchers applied fluorescent dyes to specific molecules within a cell sample. The microlenses then illuminated a smalls part of the sample with a lasers and imaged the resulting fluorescence signals. Each microlens focused on an area of about 0.6 × 0.1 mm.
The raw data produced was a series of small images, each roughly 1200 × 200 pixels wide. Individual multicolor images were then stitched together. By simultaneously imaging 13 separate color bands, a dataset consisting of nearly 16.8 billion spatial-spectral samples was produced.

Taking inspiration from modern computing methods, Orth and colleagues at Harvard and Thermo Fisher Scientific Inc. of Pittsburgh worked to overcome the limitations imposed by current multispectral microscopes. Veering away from the use of multicore processors capable of simultaneously handling massive amounts of data and instructions, Orth and his team approached this task with an imaging-based solution — microlenses.
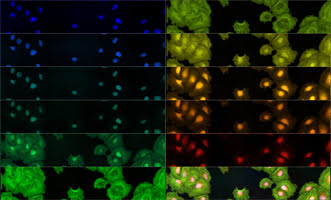
Slices of a spectral data cube. Human epithelial cells are imaged at 11 wavelengths from blue to red. The bottom right panel is a composite of all wavelength channels.
Based on single-lens designs, today's multispectral microscopes survey a single point at a time and can do so with only a few color channels (typically four or five). The process must be repeated multiple times to scan the entire sample.
The new system produces 1-MP images at a rate of 200 frames per second — much faster than what current multispectral microscopes can handle.
"What makes our microscope particularly powerful is that it records many different colors at once, allowing researchers to highlight a large number of structures in a single experiment," Orth said.
The team plans to expand its technique to produce billion-pixel, time-lapse movies of cells moving and responding to various stimuli.
The research was published in Optica (doi: 10.1364/optica.2.000654 [open access]).
Explore the 17-gigapixel image at GigaPan.