
Software interprets 3-D confocal microscopy images
David L. Shenkenberg
Neuroscience is moving from studies of a few neurons to studies of entire neural networks that more accurately show how brain cells interact and live in their natural environment. Although 3-D confocal microscopy images of the brain can provide rich information about neural networks, computers and powerful software are needed to sort through these images because they show thousands of neural features and occupy gigabytes of disk space. Researchers in New York state — at Rensselaer Polytechnic Institute in Troy and at the Wadsworth Center, a public health laboratory, in Albany — have developed Farsight, software that has demonstrated efficacy in quantifying complex brain images.
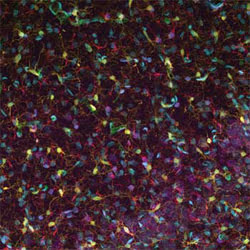
Researchers created and evaluated software for separating features from 3-D confocal images of brain slices. This image is a 2-D projection of a 3-D confocal fluorescence image of the hippocampus of a rat brain.
Co-principal investigators Badrinath Roysam and William Shain want to use the information from the 3-D confocal images to improve the lifetime of neuroprosthetic devices, which are smaller in cross-sectional area than a human hair and which typically employ microfluidics to deliver drugs or electrodes to stimulate the brain.
Neuroprosthetic devices could help patients with motor control problems and other disorders, but they usually are rejected after implantation by the natural defense mechanisms of the brain. As a result, the apparatuses cease to function after a short period. Shain said that the group uses Farsight both to quantify and to map changes in cellular organization after device insertion.
In their experiment, the researchers fluorescently labeled multiple types of cells before confocal imaging, using accepted and well-characterized labels. For example, they used the Nissl stain, which for 100 years has been used to label neurons. They captured the images using a Carl Zeiss microscope, collecting multispectral information from 32 channels. These data were unmixed into five channels showing nuclei, the Nissl stain, astrocytes, microglia and blood vessels.

This image shows the network of interactions of neural structures within the top left image. The software can quantify these interactions.
Farsight employs multiple segmentation algorithms developed over the past decade to automatically delineate all major cell types and structures in the brain and to calculate the relationships among them. The results can be inspected and validated efficiently by neuroscientists.
Last month, the researchers presented results of the software program at the Microscopy and Microanalysis annual conference. They demonstrated that the software can segment nuclei, neurons, astrocytes, microglia and blood vessels. For instance, it identified 1019 nuclei, whereas a neuroscientist looked at the same data and found 1014 nuclei, a difference of only five. Overall, the algorithms have 90 to 95 percent accuracy. Roysam said that the software is uniquely capable of conducting associative image analysis.
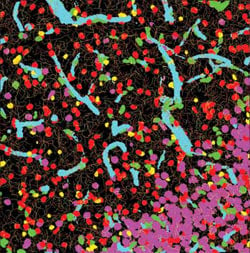
This image is a composite of automated segmentation and classification of neural structures within the left image.
Roysam said that the software not only can evaluate neuroprosthetic device implantation, but it also can quantify stem cell microenvironments. The researchers already have applied it to studying stem cell biology in controlled tissue samples, in collaboration with Sally Temple at Albany Medical College, also in New York. The scientists plan to examine neural networks throughout the brain, Shain said.
Published: September 2007