Full company details
Cobolt AB
Unit of HUBNER Photonics
 Vretenvaegen 13
Vretenvaegen 13
SE-171 54 Solna
Sweden
Phone: +46 8 545 912 30
Fax: +46 8 545 912 31
Light Sheet Microscopy Enables 3D Pathology
BioPhotonics
May/Jun 2021Adam Glaser and Jonathon Liu, University of Washington, Nicolas Reder, Light Speed Micro, Melissa Haahr, Cobolt, a part of HÜBNER Photonics
Pathology for cancer diagnosis today relies on manual processing and an individual’s assessment of 2D samples. While this method has been established for over a century, the process has limitations. It can introduce inter-observer variability, and it destroys the sample and typically analyzes under 1% of the total volume of the specimen that was removed from a patient for analysis.
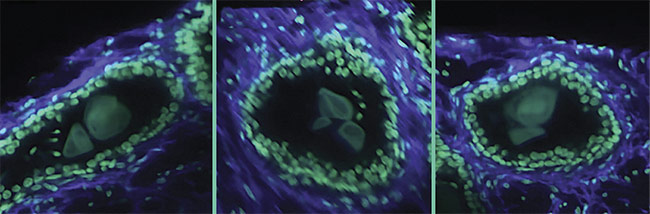
A 3D image of a human prostate biopsy. Courtesy of A. Glaser/University of Washington Molecular Biophotonics Laboratory.
Researchers are currently developing light sheet microscopy methods to overcome some of these challenges and provide a fast and effective technique that preserves 3D samples and opens the door for downstream data processing and machine learning for computer-assisted interpretation of specimens. Through innovative microscope designs and methodology, 3D pathology moves closer to clinical applications as an efficient and gentle technique for high-speed 3D imaging that can reveal important structural and molecular content in clinical specimens.
Traditional methods
Many people are familiar with and may have personally experienced the classic macroscopic medical-imaging modalities such as the CT scan, MRI, or ultrasound. A common trait of these radiology methods is that they all provide 3D macroscopic information about the body via minimally invasive methods. Radiology techniques in clinical settings largely made the transition from 2D to 3D during the 1990s. However, when it comes to analyzing samples, radiology’s microscopic sibling, pathology, has remained in a 2D world.
Radiology and pathology are often combined in the clinical diagnosis of disease, and in particular for the diagnosis and treatment of cancer. Radiology techniques provide information about the body on a macroscopic scale — for example, it can reveal the location of a suspicious lesion in the body. And pathology techniques, in particular histology, are used to provide microscopic information about tissues and cells from extracted samples, such as a tissue biopsy or a blood sample, to better understand the progression of a disease.
In pathology, tissues are prepared and interpreted according to dated protocols (Figure 1). Typically, the procedure begins with the removal of a sample of tissue from a patient to screen for disease (for example, cancer) or another medical condition. The specimens are then stabilized using harsh fixatives and dehydrated using alcohols such as xylene and ethanol, so that the specimens can ultimately be embedded in paraffin wax blocks. The wax blocks are then mounted on microtomes so that thin sections of several microns each can be cut from the face of the block. The slices are mounted on a glass slide and stained with chromogens — most commonly hematoxylin and eosin (H&E). The stained slides are then manually interpreted by a pathologist on a traditional analog microscope.
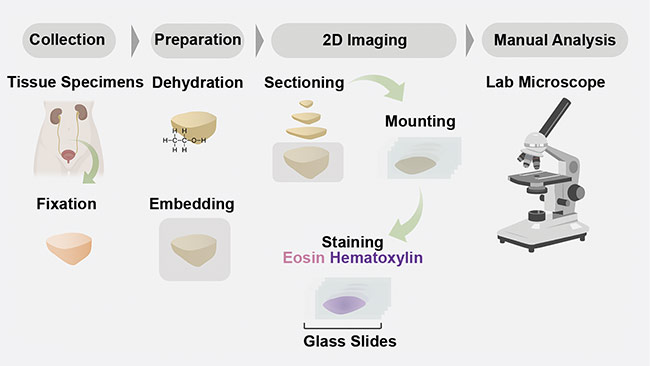
Figure 1. A schematic of a conventional 2D formalin-fixed paraffin-embedded hematoxylin and eosin
(H&E) pathology workflow. Courtesy of A. Glaser/University of Washington Molecular Biophotonics
Laboratory.
While this workflow has been the standard for over a century, it cannot provide a full diagnosis of a condition or illness. Because only a small number of glass slides are prepared from each specimen, the reliability of current pathological tests may suffer from undersampling, meaning a critical part of the specimen may be missed, or information related to disease progression may not be observed.
For example, a single 5-µm-thick section of a 1-mm-thick tissue specimen represents only 0.5% of the total tissue. In addition, the process of preparing glass slides in this workflow is time-consuming and destructive, which limits compatibility with emerging molecular medicine assays for additional analysis of the same sample. Finally, the glass slides are typically assessed manually using an analog light microscope. Therefore, digital data is only available after using a digital whole-slide scanner that converts data from the slide into a high-resolution digital image. While these scanners have recently gained FDA approval, they add an additional time-consuming step to the already laborious pathology workflow.
Therefore, there is a need and the potential for a more efficient pathology workflow that comprehensively samples specimens and provides digital imaging data without compromising the specimen quality.
Importance of 3D imaging
In addition to the limited sampling of a specimen, current pathology methods provide only a 2D view of specimens that are naturally 3D. This can be troublesome for analyzing certain biological structures deep within a sample that may hold valuable information relevant for diagnosis and treatment.
For example, convoluted structures such as vessels, lymphatics, and glands often resemble tree-like frameworks in 3D that cannot be accurately captured at depth in 2D. These structures are of interest to analyze because their condition can be predictive of several diseases, and therefore accurate interpretation is of paramount importance.
Complex distributions of various cell types are also of interest — for example, in the tumor immune environment, where various cellular processes in effect protect the tumor from the immune system and allow for metastasis. The spatial relationships between various immune cell types may be predictive of treatment response in certain patient populations, but these relationships can be difficult to quantify accurately in 2D. Finally, rare cell types, such as those that may initiate and promote metastatic spread of cancers, may be missed entirely due to the limited sampling capabilities of the current pathology workflow (Figure 2).
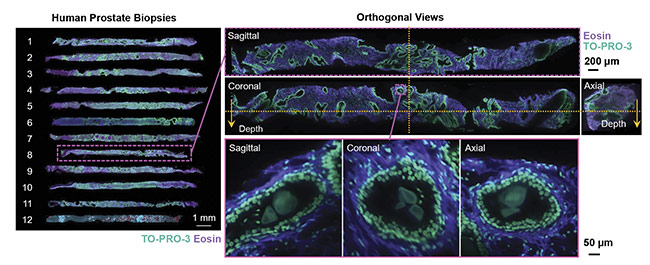
Figure 2. 3D images of human prostate biopsies demonstrating the type of cellular information available from 3D sample analysis that is not available from standard 2D histology slides. Courtesy of A. Glaser/University of Washington Molecular Biophotonics Laboratory.
Along with the additional biological information available from 3D imaging, specimen handling is also improved in a 3D imaging protocol versus the current 2D workflow. For example, the specimen is better preserved because the sample is not sliced, which can allow for multiple tests to be performed on the same specimen. Also, the 3D imaging protocols will assist in digitalizing the data collected (Figure 3). This is important because the assessment of pathology samples is the primary tool for diagnosing cancer and determining the best treatment plan for a patient. The improved preservation of samples and digitalization of the image data could help to improve diagnosis and treatment plans for patients, as well as aid doctors and pathologists in making critical decisions, via machine learning and automated image analysis.
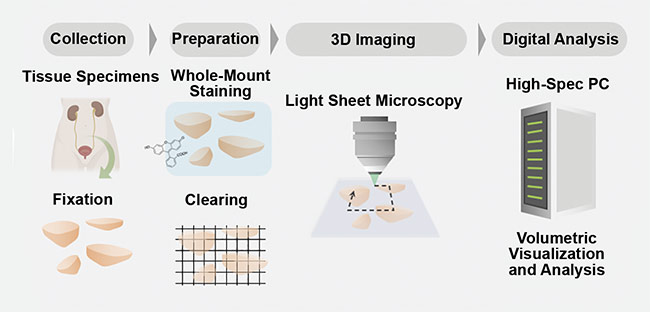
Figure 3. A schematic of a comprehensive 3D pathology workflow. Tissue samples are collected from a patient and fixated. Then the samples undergo a tissue-clearing procedure and are imaged in 3D with digital analysis. Courtesy of A. Glaser/University of Washington Molecular Biophotonics Laboratory.
Light sheet microscopy’s role
Despite the benefits of analyzing pathology samples in 3D, there are still challenges in developing the technology and methodology that can replace current pathology techniques. New techniques must be reliable and have a high enough throughput for clinical applications. Light sheet microscopy has emerged as a viable technology to address these challenges.
Historically, other technologies that theoretically enable 3D pathology have been pursued by scientists and manufacturers. However, previous approaches were lacking logistical feasibility due to their complexity in a laboratory, and throughput was not high enough for routine clinical use. The techniques were limited by the complex microscopy methods required for imaging biological tissues, which are naturally opaque. The methods proved to be too slow for imaging clinical tissues within a reasonable timeframe. However, the development of highly effective optical clearing techniques has opened the door to faster, more optically efficient techniques.
In combination with these new clearing methods, light sheet microscopy has emerged as the fastest and most efficient imaging method with clinical possibilities for 3D pathology.
Unlike traditional fluorescence microscopy methods, which often use a single objective lens and light path to simultaneously illuminate and image a specimen, light sheet microscopy uses two objective lenses and separates the illumination and collection paths of the microscope. By placing these two objective lenses at 90°, a thin sheet of light can be used to optically rather than physically section a specimen. Coplanar alignment of the light sheet to the collection objective’s focal plane provides selective illumination within the specimen. This is in contrast to traditional fluorescence microscopy methods, in which the entire sample is often illuminated to collect information from just a single point or plane within the specimen (Figure 4).
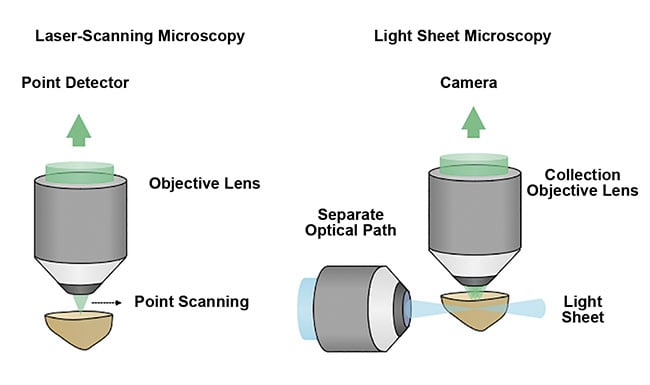
Figure 4. A schematic comparing point- or laser-scanning microscopy with light sheet microscopy
techniques. The optical sectioning of a sample by light sheet microscopy is a more efficient and gentle method that enables high-speed 3D imaging with minimal damage to the sample. Courtesy of A. Glaser/
University of Washington Molecular Biophotonics Laboratory.
When combined with tissue clearing, light sheet microscopy can provide unprecedented access to the structural and molecular content in clinical specimens.
Open-top light-sheet microscopy
In their early development, light sheet microscopes were purposefully designed around the analysis of a single live specimen. But in recent years, the advantages of light sheet microscopy have also been harnessed for high-speed 3D imaging of larger clinical specimens
1. Unfortunately, light sheet systems that were initially designed for live specimens are not optimal for clinical specimens. For example, the orthogonal arrangement of the light sheet objective lenses have imposed constraints, limiting the size, shape, and/or number of specimens.
To address this limitation, researchers at the University of Washington Molecular Biophotonics Laboratory — led by professor Jonathan Lui, with Adam Glaser and Nicholas Reder — have designed open-top light-sheet (OTLS) microscopes that are optimized for 3D clinical specimens
1. The design overcomes the spatial constraints of traditional systems by placing all optical components below the specimen. This allows the microscope to accommodate specimens of arbitrary size without physically interfering with the illumination and collection objective lenses (Figure 5).
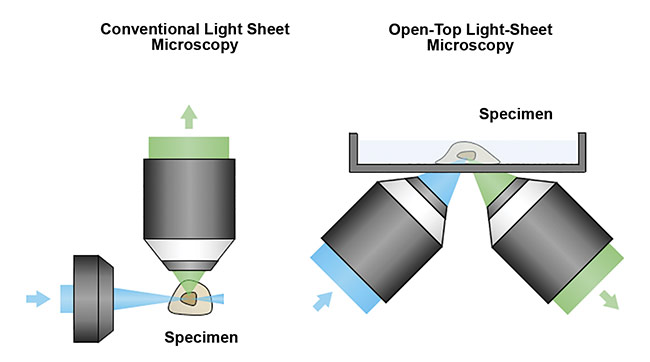
Figure 5. A schematic of a conventional light sheet microscope (left) and an open-top light-sheet microscope (right). Unlike the conventional design, the open-top geometry enables simple mounting of specimens on top of the system, serving as a flatbed scanner for tissues. In addition, the angled objectives place no constraints on the lateral dimensions of the specimen, which is critical for clinical applications in which tissues often have large lateral dimensions. Courtesy of A. Glaser/University of Washington Molecular Biophotonics Laboratory.
However, this geometry also presents unique optical challenges because the off-axis (45°) illumination and collection beams are not easily index-matched into the sample, which can result in aberrations. To enable aberration-free imaging, the group developed innovative optical solutions, including the use of a solid immersion lens (SIL), a solid immersion meniscus lens (SIMlens), and customized multi-immersion objective lenses
2.
Unlike confocal or multiphoton-based systems, the OTLS device is relatively simple to use and contains no mechanically moving parts other than a standard galvanometric scanning mirror and a motorized xyz sample stage. Without the need for high-speed resonant scanning mirrors and/or complicated pulsed laser sources, operators of the OTLS system can avoid common optomechanical failures caused by too many moving parts (Figure 6).
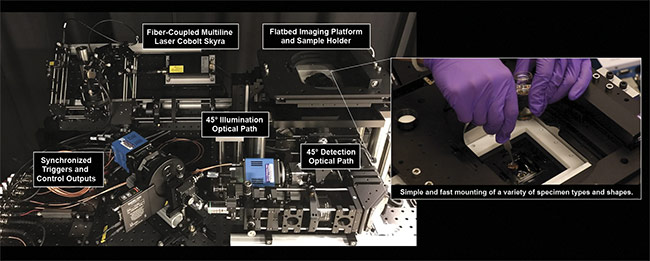
Figure 6. The University of Washington Molecular Biophotonics Laboratory’s open-top light-sheet microscope. It features 45° illumination and collection
paths, and a flatbed sample platform for mounting samples of various shapes and sizes. Courtesy of A. Glaser/University of Washington Molecular
Biophotonics Laboratory.
Unlike other 3D-imaging approaches, which are either too slow or too complicated for routine clinical use, the OTLS devices are fast and easy to use, providing submicron imaging at a rate of ~1 mm
3/min
2. In addition, multiple specimens can be mounted simultaneously on the microscope, making it compatible with high-throughput imaging, which is critical for the success of any technology being pursued for routine clinical use.
Simple clearing protocol
Although many optical clearing protocols exist, the requirements for clinical pathology are unique in several ways. The protocol must be simple and fast, and ideally it will not require the use of toxic reagents. And while many preclinical applications use tissues with endogenous fluorescent proteins, which are often quenched during the clearing process, clinical human tissues do not contain fluorescent proteins. Therefore, the group adopted a simple and straightforward solvent-based clearing protocol that uses food-grade cinnamon oil that renders millimeter-size tissues transparent in under 24 hours.
The fluorescent labeling protocol has been developed, along with the OTLS microscope, to mimic the appearance of the chromogenic H&E stains that are used in current pathology samples. To re-create the appearance of hematoxylin, a nuclear stain, the red fluorescent dye TO-PRO-3 Iodide is used. Eosin itself is a green fluorescent dye and can be used directly in the protocol.
Both of these fluorescent channels are inherently grayscale in digital images. Therefore, to reproduce the pink and purple appearance of H&E images, digital false-coloring is used to mimic the H&E appearance for analysis (Figure 7).
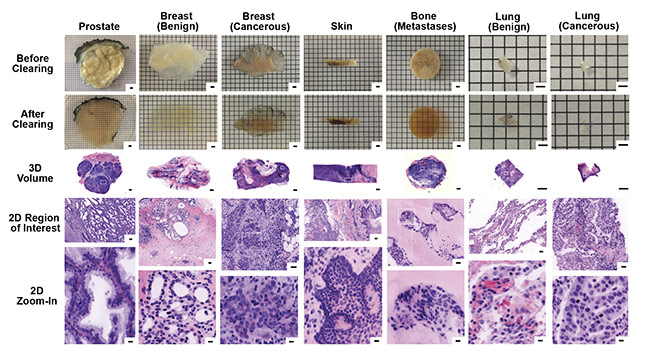
Figure 7. Images of 3D samples with digital false coloring to mimic the appearance of H&E stains of traditional 2D samples. The digital 3D images allow for
2D zoom-in on regions of particular interest, with higher resolution than traditional 2D slide sample analysis. Samples include biopsies of prostate, breast (benign and cancerous), skin, bone (metastases), and lung (benign and cancerous). Scale bars: 2 mm (first three rows, 3D), 100 µm (fourth row, 2D region of interest), and 10 µm (bottom row, 2D zoom-in). Courtesy of A. Glaser/University of Washington Molecular Biophotonics Laboratory.
Compact, reliable hardware
Although the dyes mentioned above can be excited by lasers in the red and green portions of the spectrum, certain applications require additional multiplexing. Therefore, it is advantageous to have an excitation source with four channels spanning the visible spectrum (for example, 405, 488, 561, and 638 nm) to enable simultaneous analyses and expanded functionality of the microscope. And, due to the inherent efficiency and gentle exposures of light sheet microscopes, the excitation lasers required are of moderate output power (50 to 100 mW per wavelength), and modulation controls for synchronization are at frequencies often <1 kHz. This opens up the possibility of using compact and permanently aligned multiline laser solutions to increase excitation color capabilities without adding system complexity.
Multiline lasers meet microscopy
In the case of Cobolt’s multiline laser, the Cobolt Skyra, all the miniaturized optical elements of the laser sources and the combining optics are precision aligned and permanently fixed onto thermally controlled platforms in hermetically sealed packages. The optical elements are fixed using a proprietary technique, HTCure, which is based on careful
thermomechanical matching and high-
temperature curing that leads to very strong fixation. In this way, the permanently aligned output beam can be coupled into a single-mode polarization-maintaining fiber with stable output power, even in varying temperatures or operating conditions.
A multiline laser concept enables smaller and more cost-efficient multicolor bioimaging instrumentation that is easier to manufacture and maintain. This supports the drive to bring more advanced laser-based microscopy techniques into clinical settings for improved medical diagnostics and the further development of new analytical techniques, including a 3D pathology solution.
View for the future
The transition from 2D to 3D pathology has the potential to improve the diagnosis and treatment of cancer. With advancements in methodology (such as new clearing techniques) and hardware (such as multiline lasers), as well as unique and robust microscope designs (such as the open-top light-sheet microscope), light sheet microscopy has emerged as the fastest and most efficient imaging method with clinical possibilities for 3D pathology.
The advantages of 3D pathology include increasing the total sampling volume, analyzing intact tissue and 3D structures of importance, digitalizing the data collection, maintaining the sample integrity for additional analysis, and reducing the dependence on manual processing and interpretation of sample images.
Remaining challenges include processing time and clinical acceptance. The open-top light-sheet microscopy technique overcomes the typical challenges of clinical 3D pathology due to its ease of use, robust design, ability to image samples of various sizes and shapes, and safe sample-clearing method.
New developments in light sheet microscopy offer viable solutions for 3D pathology in clinical applications. The acceptance of these new 3D pathology methods will depend on large-scale clinical studies that quantitatively demonstrate the advantage of 3D pathology methods over the current 2D pathology workflow. These studies are currently underway at the University of Washington Molecular Biophotonics Laboratory and elsewhere.
Meet the authors
Adam Glaser is an instructor in mechanical engineering at the University of Washington. He received a B.S. in mechanical engineering from Tufts University in 2010 and a doctorate in engineering sciences from Dartmouth College in 2015. He completed his most recent postdoctoral research at the University of Washington in 2019. Working with Jonathan Liu, Glaser helped to establish the emerging field of 3D pathology. His graduate and postdoctoral research has been featured by numerous media outlets and has led to the co-founding of two funded startup companies, DoseOptics and Lightspeed Microscopy; email:
[email protected].
Melissa Haahr is a product manager at Cobolt AB, part of HÜBNER Photonics, in Stockholm. She received a B.A. in chemistry and environmental science from Alfred University in 2011, and an M.S. in physical chemistry from the University of Oregon in 2012. Haahr joined Cobolt in 2014; email:
[email protected].
Acknowledgments
The authors appreciate the work of the following people in the production of this article: Nicholas Reder, M.D., CEO of Lightspeed Microscopy, and Jonathan Liu, professor at the University of Washington and director of its Molecular Biophotonics Laboratory.
References
1. A. Glaser et al. (2017). Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens.
Nat Biomed Eng, Vol. 1, Issue 7, Article No. 0084.
2. A. Glaser et al. (2019). Multi-immersion open-top light-sheet microscope for high-throughput imaging of cleared tissue.
Nat Commun, Vol. 10, Article No. 2781.