High-speed imaging reveals information about how the Golgi apparatus works.
Hank Hogan, Contributing Editor
Within
cells, the Golgi apparatus gets things where they need to go. An organelle found
in all eukaryotic organisms, the Golgi apparatus primarily takes in protein-containing
vesicles, then modifies, sorts and ships them off to their final destination. The
exact process by which this happens, however, has been in dispute.
Now, thanks to advances in image-capture technology,
improvements in image processing and some careful research, two groups — one
in Japan and another in the US — have clearly seen what is going on inside
the Golgi apparatus. Their results indicate that transport containers within the
complex mature along with the developing biochemicals from an initial to a final
form.
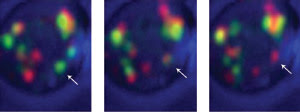
Labeling early Golgi cisternae with GFP and late cisternae with DsRed and using a high-speed 4-D
confocal microscopy system, investigators observed that individual cisterna (arrow)
changed from green to yellow to red, indicating that cisternae were maturing from
an early to late stage and not acting as constant compartments. Courtesy of Benjamin
Glick, University of Chicago.
The fast 4-D fluorescence microscopy
technology employed in these investigations could be used to tackle other cell biology
problems because at least one version is transitioning from something custom built
to an off-the-shelf product. “This system will be commercialized soon. Indeed,
the companies involved in this project plan to do so in a few years,” said
Akihiko Nakano, a professor of biological science at the University of Tokyo.
Nakano, who also is director of the
molecular membrane biology laboratory at the Riken Discovery Research Institute
of Saitama, led the Japanese team. Besides investigators from the university and
from Riken, the group included a researcher from the Yokogawa Electric Corp.’s
Biocenter.
Spinning disks and yeast
Although their results agree, the two groups reached
their conclusions via different paths. The team in Japan used a custom-designed
high-performance laser scanning microscopy system that combined spinning-disk confocal
scanning with a high-gain avalanche rushing amorphous photoconductor camera. This
system, according to Nakano, was critical to the success of the experiment.
Using a custom spinning-disk confocal and fast camera microscopy
setup, researchers showed that Golgi cisternae matured from an early to late stage
while transporting proteins. The team achieved 50-nm resolution in three dimensions,
while still being able to image rapidly enough to track the maturation process.
Courtesy of Akihiko Nakano, University of Tokyo and Riken Discovery Research Institute.
Unlike a laser-scanning confocal microscope,
a spinning-disk confocal system does not scan a laser across a specimen, which can
take seconds. Instead, a spinning pinhole array quickly sweeps the beam across an
area. When combined with a microlens array, the result is a scan time measured in
milliseconds. The researchers combined this approach with a high-gain avalanche
rushing photoconductor camera, which takes advantage of the photoconduction that
occurs in bulk amorphous selenium. The resulting amplification makes weak signals
more easily detectable.
Nakano noted that combining the technologies
was not very difficult but that pushing the performance was challenging. However,
the researchers succeeded in hitting a number of technological high points. “The
frame rate in two dimensions can be 180 per second, and the piezo actuator system
can scan 100 μm in 0.1 second in the Z-axis,” he said.
In the past, it was not clear exactly
how transport takes place with the Golgi complex. Endoplasmic reticulum proteins
enter the complex in a compartment called the cis cisterna — the first
of the three main plates that comprise the Golgi apparatus. The proteins then undergo
modifications and exit from the last, or trans, cisterna. One hypothesis
was that transport takes place in unchanging cisternal compartments, while another
postulated that the cisternae are transient, being newly created each time, maturing
from cis to trans and then disappearing.

Endoplasmic reticulum proteins enter the Golgi apparatus in an early
compartment called the cis cisterna, where they undergo modifications, and exit
from a late, or trans, cisterna. The researchers found that cis cisternae mature
into trans cisternae, denoted here by red to green fluorescence. Courtesy of Akihiko
Nakano.
As detailed in the June 22 issue of
Nature, the researchers studied the yeast Saccharomyces cerevisiae,
largely because the Golgi apparatus within these organisms consists of a single
layer. Thus, there was no need to try to unravel images from a stacked structure,
which is found in many animal and higher plant cells.
They fluorescently labeled biomolecules,
choosing some that resided mainly in the cis and others found in the trans.
They fused a GFP molecule to both groups while they fused a red fluorescent protein
to the complementary biomolecule. In this way, some cis markers fluoresced
green and some trans fluoresced red, as well as vice versa. They carefully
selected these constructs, Nakano said, because the fused proteins were biologically
active and did not interfere with the functioning of the Golgi apparatus.
They then tracked what happened in
living yeast cells. They looked at the color of individual cisterna in real time,
using high-speed imaging to follow the compartment. In one case, cisternae would
be green at the beginning and turn red over the course of 3 min. In the other labeling
situation, the cisternae would start out red and turn green within a few minutes.
No matter how the labeling was done, the reverse transformation never happened.
These results indicated that the cisternae
were not passing the proteins along from one compartment to another. Instead, they
showed that the cisternae matured from early to late stage — from cis
to trans. Nakano said that he favored the maturation model from the beginning
but that seeing the visual proof was electrifying. “When I first saw the color
change, I was really excited,” he said.
The scientists are working to improve
the sensitivity of the camera and the overall system. The next step will be to try
to understand the molecular mechanism of cisternal maturation, which their findings
indicate involves dynamic membrane mixing and segregation. They also are looking
into using various yeast mutants, which can be challenging because many of them
are sensitive to temperature. Thus, temperature must be changed during microscopic
observation.
The US investigators — from the
University of Chicago and headed by Benjamin Glick, an associate professor of molecular
genetics and cell biology — also worked with a custom-built confocal microscopy
system.
To speed up the scanning process, they
devised a continuous-movement method for the Z-axis. This worked, Glick noted, because
it eliminated delays between successive optical sections without compromising image
quality. With an average velocity of 3.5 μm/s, the movement of the objective
during the 1.67 ms needed for a line scan was only 6 nm, too small a distance to
have an effect.
Because they needed to image a cell
for several minutes, the researchers had to avoid photobleaching. Therefore, they
had to scan as fast as possible at the limit of resolution with the laser power
turned down as much as possible. Glick said that, besides image capture, image processing
was essential because of the data created under these conditions. “The resulting
images are noisy,” he said.
In their investigations of Golgi maturation,
the scientists used a multistep scheme to improve the images. They first ran the
slices through a 3 x 3 hybrid median filter, which removed shot noise arising from
statistical fluctuations without degrading the signal. They sharpened the image
using iterative deconvolution via software from the Netherlands-based Scientific
Volume Imaging BV, then followed that by average projecting the images using NIH
Image, a software package from the National Institutes of Health, and by adjusting
contrast and subtracting background. Finally, they used Photoshop to merge captured
fluorescence images with scattered light images of the cells for a visual presentation.
They tracked individual cisterna within
S. cerevisiae by investigating Golgi proteins that marked the appropriate
compartments and that were abundant or capable of being overexpressed without interfering
with the organization or function of the complex.
They tried many candidates before finding
two — Vrg4 and Sec7 — that worked. They also developed a monomeric variant
of the fluorophore DsRed, linking multiple monomers in tandem to generate the strong
signal required by the imaging conditions. “Strong fluorescence signals were
needed so that we could image at very low laser excitation levels,” Glick
said.
They tagged Vrg4 with a GFP molecule
and Sec7 with their DsRed variant and then followed many individual cisterna in
real time. They found that the cisternae started out green, with the fluorescence
peaking shortly after acquiring the labeled Vrg4. The intensity then declined and
the cisternae became red over the span of many minutes. There were no cases where
the reverse happened. Other measurements showed that the cisternal maturation was
fast enough to account for the observed kinetics of transport through the secretory
pathway.
Clear-cut results
Glick noted that these results were not unexpected,
but he was surprised that they were so clear-cut. There was little fusion or fission
of cisternae, making it easy to follow individual ones for long periods. The research
also was published in the June 22 issue of Nature.
The researchers are working with another
group to complement their fluorescence microscopy with electron tomography to produce
high-resolution 3-D structural data. They also are looking at using fluorescently
tagged yeast mutants, which will have specific and known secretory path defects,
in an effort to determine which proteins are crucial for Golgi maturation.
Finally, they are considering verification
of a basic prediction of the maturation model, which conjectures that a secretory
cargo will remain associated with a cisterna while resident Golgi proteins come
and go. Doing that, however, will require fluorescently tagging a secretory cargo
as well as resident Golgi proteins. It won’t be easy to do, Glick remarked.
“The technical challenges are
to develop a suitable fluorescent secretory cargo for yeast, and to perform three-color
4-D imaging to observe this cargo together with two Golgi markers,” he said.